- Homepage
- Key Information
- Students
- Staff
- PGR
- Health and Safety
- Computer Support
- National Student Survey (NSS)
- Intranet Help
Prof Edward Keedwell
Research
Publications
Prof. Keedwell has published one book 'Intelligent Bioinformatics' and over 120 papers in journals and conferences in the fields of computer science, bioinformatics and hydroinformatics. Details of these are available on the publications page. Further details are also available on Google Scholar and Researchgate
Funded Projects
Prof. Keedwell has successfully applied for projects worth over £2m in total. This funding has predominantly been provided by the EPSRC, with projects also funded by Innovate UK (was the Technology Strategy Board) and by industry.
Previous/Current Project Websites
Ant Colony Optimisation for Genome-Wide Association Studies (ACOGWAS)
This EPSRC funded project is investigating the use of a modern nature-inspired optimisation technique known as ant colony optimisation to discover associations between groups of small changes in DNA and a variety of diseases including type I and type II diabetes, inflammatory bowel disease and rheumatoid arthritis. The developed techniques are designed to search the billions of combinations of small changes in DNA over populations of thousands of people to determine those that might lead to a greater susceptability to disease.
In many cases, it is not possible to search every combination, even with very powerful computers and so this project is using one of a family of stochastic optimisation algorithms to perform this task. You can read more about the biology and computer science of this project, as well as read about the progress we are making in the sections below.
Ant Colony Optimisation (ACO)
The first ACO algorithm, proposed by Marco Dorigo in 1996 [1] was Ant System and since then a number of modifications have been proposed to improve performance of the algorithm. ACO is inspired by the way that populations of ants in nature can find a short path between the nest and a food source by sensing the pheromone trails of previous ants. The computational algorithm requires ants to traverse a topology which is either a direct representation of the problem (e.g. in the travelling salesman problem) or a landscape of variable choices known as a construction graph. We have adapted the algorithm to work with the specific challenges of Genome-Wide Association Studies.
Subset-Based ACO
Standard ACO works by laying pheromone on the links between variables choices, which works well for combinatorial problems. However, for subset based problems (i.e. selecting a small number of N from a larger set), this method does not work well and incurs high computational complexity. We have therefore made use of a method first proposed by Leguizamon and Michaelawicz (1999) [2] which lays pheromone on the variables (SNPs) themselves rather than the links between them, reducing the search space considerably and speeding execution.
Tournament Selection ACO (T-ACO)
Standard ACO path selection works by constructing a roulette wheel of variable choices for the ants to consider, weighted by the pheromone (and possibly a local heuristic) on each choice. This works well for a wide range of problems, but the specifics of GWAS mean that such a roulette wheel would require ~400,000 such segments, leading to poor path selection pressure and large computational loads. To counter this, we developed a tournament-based ACO algorithm [3] that chooses paths by randomly selecting N SNPs and choosing the SNP with the best pheromone value for the ant to explore. Experimentation has shown the effectiveness of this method on high-dimensional problems and improved execution times for reasonable N.
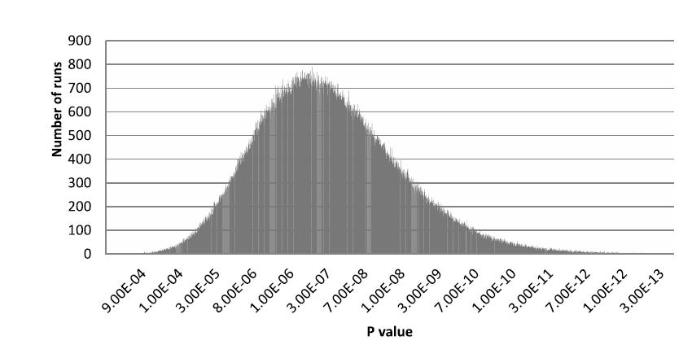
P-Values of randomly generated gene-gene interactions in Type-II diabetes data.
GWAS Using ACO
We have applied the above methods to data from the Wellcome Trust Case Control Consortium involving a number of different diseases. The datasets consist of around 400,000 SNPs and over 5000 individuals (~3000 controls, ~2000 cases) that must be explored by the ACO approaches. The size of this dataset has presented some challenges from a computational perspective and the flexibility of the ACO approach is such that we have been able to explore interesting relationships between SNPs. These are presented in more detail below:
Byte-Level Data Compression
With between 400,000 and 500,000 SNPs and 5000 individuals, storing the datasets in working memory became a problem. Using a single byte for each SNP would require between 2 and 2.5 GB of free space in RAM which can result in the OS paging to disk even with 4 GB or more of memory. We have therefore developed a compression technique that allows 4 SNPs to be stored in a single byte that reduces runtimes and allows for multiple runs to be made in parallel on the same machine if required [4].
Logical Operations
The traditional view of gene-gene interactions is that they operate with an AND operation, for instance - SNP1=GENOTYPE X AND SNP2=GENOTYPE Y is commonly used. We have explored some of the relationships that might exist between SNPs that are not represented by this AND operation, and have included OR, XOR and NOT to provide the algorithm more flexibility in the way in which it allows SNPs to interact [5]. Results have shown that these additional operations provide additional power to the algorithm to find statistically important associations in the data.
Assessing Statistical Significance
A key issue with developing these methods is the correction for multiple testing. With single associations, it is relatively simple to correct p-values using Bonferroni correction, but a stochastic algorithm discovering gene-gene interactions is more difficult. We have used monte-carlo methods to select 1,000,000 random combinations from the data and have also run the algorithm thousands of time on shuffled data to determine the statistical significance of results.
Summary
This ongoing EPSRC project has yielded interesting developments in ACO, the representation of data and significance testing and has shown promise in discovering gene-gene interactions in large-scale GWAS data.
References
- M. Dorigo, V. Maniezzo, et A. Colorni (1996) Ant system: optimization by a colony of cooperating agents, IEEE Transactions on Systems, Man, and Cybernetics--Part B , volume 26, numéro 1, pages 29-41
- G. Leguizamon and Z. Michalewicz, (1999) "A new version of ant system for subset problems," Proceedings of the 1999 Congress on Evolutionary Computation(CEC 99), vol.2, pp.1458-1464, 1999.
- Sapin E, Keedwell EC. (2012) "T-ACO - Tournament Ant Colony Optimisation for High Dimensional Problems", ECTA 2012 - 4th International Conference on Evolutionary Computation Theory and Applications, Barcelona, Spain, 5th - 7th Oct 2012.
- Sapin E, Keedwell EC., Frayling T (2013) "Subset-Based ACO for Genome Wide Association Study: Discovery of Promising Combinations" to appear at Genetic and Evolutionary Computing Conference GECCO 2013
- Sapin E, Keedwell EC., Frayling T (2013) "Ant Colony Optimisation for Exploring Logical Gene-Gene Associations in Genome Wide Association Studies", International Work Conference on Bioinformatics (IWBBIO), Granada, Spain.
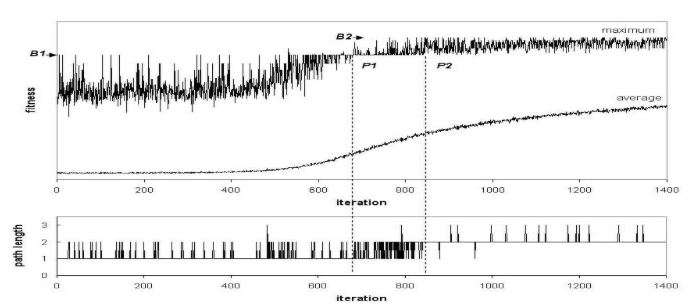
Pheromone trail for initial ACO algorithm over time note the inflection point B2
Sequence Analysis Based Hyperheuristics for Real World Problems (SEQAH)
Hyperheuristics are a set of techniques that optimise at the level above metaheuristics (such as evolutionary algorithms and particle swarm optimisation). They can be used to discover near-optimal combinations of low level heuristics (e.g. mutation and crossover operations) and are known as selection hyperheuristics or can generate new heuristics to aid in solving difficult problems and are known as generation hyperheuristics. We have developed new methods for both types of hyperheuristics and have applied the resulting algorithms to a wide variety of problems, including those in the water industry.
Sequence Analysis Based Hyperheuristics for Real-World Problems
This EPSRC-funded project is investigating the use of sequence analysis techniques taken from bioinformatics in the development of hyperheuristics. Hyper-heuristics are optimisation approaches that operate at the level above traditional metaheuristics (e.g. evolutionary algorithms) and must either select from a pre-defined set of low level heuristics (e.g. mutation, crossover and hillclimbing operators) or generate them. The focus in the SEQAH project is on developing selection hyperheuristics by analysing and creating sequences of heuristic usage during an optimisation. The first method developed in this project is known as SSHH and uses a hidden markov model to analyse and produce sequences of heuristics.
Sequence Based Selection Hyperheuristic (SSHH)
SSHH uses a hidden markov model where hidden states are replaced with low level heuristics (LLH) and a matrix of transition probabilities determine the movement between these states. A further set of emission probabilities determine whether, given a particular LLH selection, the heuristic will be applied or will be coupled with another LLH to form a sequence. The method can be seen in the diagram below and more detail can be found in this paper (best paper nominee, GECCO 2015).
Promising early results obtained with minimal tuning or problem specific expertise:
- SSHH obtains better performance than the winner of the Cross Domain Heuristic Challenge (CHESC) 2011
- Ranked 3rd in the Windfarm Optimisation Challenge at GECCO 2015
- Outperformed genetic algorithm formulations on the water distribution network optimisation problem
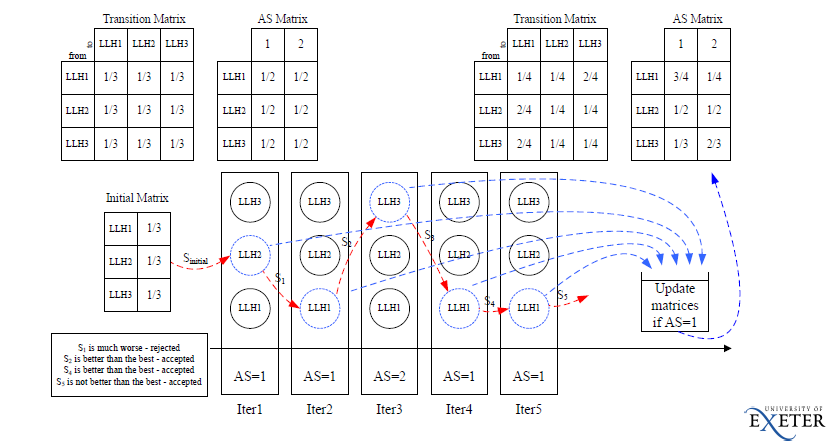 SSHH Operation. AS= Acceptance Strategy (1=evaluate new solution, 2=move to next LLH), LLH = Low Level Heuristic
SSHH Operation. AS= Acceptance Strategy (1=evaluate new solution, 2=move to next LLH), LLH = Low Level Heuristic
Multi-objective hyperheuristics and discolouration propensity modelling
In this project, we are investigating new hyperheuristics for continuous & mixed encoding multi-objective problems and are applying them to problems in the water industry, including the mitigation of discolouration risk. The project is supported by EPSRC and consultants Mouchel Ltd.
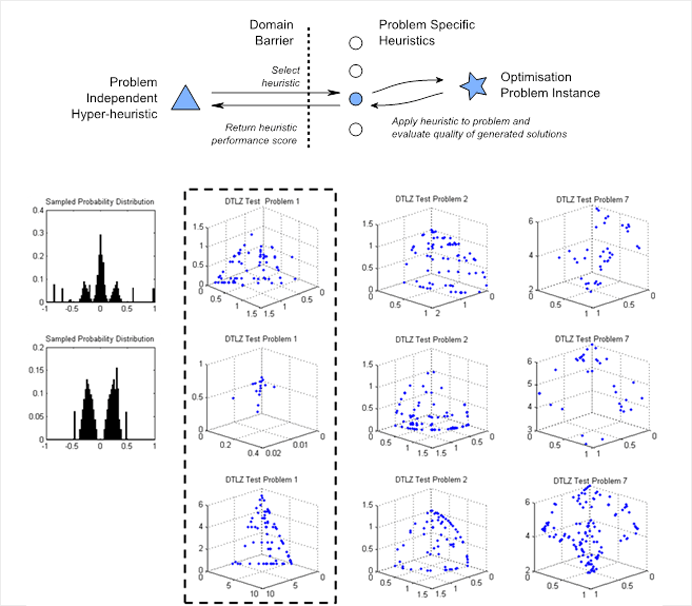
Heuristics evolved for solving multi-objective water distribution network problems with discolouration risk taken from the pareto-front of operations. The probability distributions show the probability of mutation around the current point and the point clouds demonstrate the resulting behaviours in optimisation space.
- Kheiri A., Keedwell E. (2015) Markov Chain Selection Hyper-heuristic for the Optimisation of Constrained Magic Squares accepted to UKCI 2015, Exeter, UK
- Kheiri A., Keedwell E., Gibson, M., Savic, D., (2015) Sequence analysis-based hyper-heuristics for water distribution network optimisation accepted to Computing and Control in the Water Industry (CCWI) 2015, Leicester, UK
- Kheiri, A., Keedwell E., (2015) A sequence-based selection hyper-heuristic utilising a hidden Markov model in the proceedings of Genetic and Evolutionary Computation Conference (GECCO) 2015, Madrid, Spain, p417-424 (best paper nominee)
- McClymont K, Keedwell E, Savic D. (2015) An analysis of the interface between evolutionary algorithm operators and problem features for water resources problems. A case study in water distribution network design, Environmental Modelling and Software, DOI:10.1016/j.envsoft.2014.12.023
- McClymont K., Keedwell E., (2011) “Deductive Sort and Climbing Sort: New Methods for Non-Dominated Sorting” Evolutionary Computation 2011, MIT Press
- McClymont K., Keedwell E., (2011) “Benchmark Multi-objective Optimisation Test Problems with Mixed Encodings” IEEE Congress on Evolutionary Computation 2011, New Orleans
- McClymont K., Keedwell E., (2011) “Markov chain Hyper-heuristic (MCHH): an Online Selective Hyper-heuristic for Multi-objective Continuous Problems” Genetic and Evolutionary Computing Conference 2011, Dublin, Ireland
- McClymont K., Keedwell E., (2010) “Optimising Multi-Modal Polynomial Mutation Operators for Multi-Objective Problem Classes” IEEE Congress on Evolutionary Computation 2010, Barcelona
